Redefining Spatial Transcriptomics: M20 Spatial
In August 2022, we proudly introduced M20 Seq®, our revolutionary single-cell sequencing technology based on random primers, setting a new standard in comprehensive transcriptome analysis. Accompanying this innovation was the launch of our VITA Single-Cell Full-Length Transcriptome Sequencing Platform, enabling the comprehensive exploration of whole transcriptomes across diverse sample types and species.
Our dedication to scientific progress extends beyond existing technologies. Acknowledging the intricate multidimensional complexities of biological mechanisms, which span both the temporal and spatial scale, we are inspired to foster novel and comprehensive insights into their complex organization and regulation. This commitment fuels our continuous efforts to deliver innovative approaches that push the boundaries of current technologies, paving the way for pioneering discoveries that will shape the future of biomedicine.
We are thrilled to present a groundbreaking advancement: 'M20 Spatial' – our revolutionary spatial whole transcriptome solution, powered by our random-primer technology. M20 Spatial marks a groundbreaking shift in spatial transcriptomics, expanding species applicability and sample type compatability, improving RNA capture efficiency, and enhancing sequence coverage. With M20 Spatial, we empower the scientific community to delve deeper into the spatial and temporal dimensions of cellular processes, driving advancements and leading discoveries at the forefront of biomedical research.
Embrace the next generation of spatial transcriptome technology with M20 Spatial!
A New Dimension with Spatial Transcriptomics
Pioneering significant advancements in life sciences and medicine, spatial transcriptomics was recognized as the "Technology of the Year" by Nature Methods in 2020. Since then, its application in scientific research has experienced exponential growth, as evidenced by an increase in publications employing this technology (Figure 1). The pace of progress in the areas of application and industrial implementation has notably accelerated. These developments are significantly contributing to decipher key questions of cell biology, developmental biology, neuroscience, oncology and more.

Figure 1: Number of articles employing spatial transcriptomics on Pubmed in recent years
In contrast to conventional single-cell transcriptomics, spatial transcriptomics provides the possibility to correlate gene expression with precise spatial locations. This additional dimension enables a comprehensive understanding of the spatial distribution of diverse cell types and states, facilitating precise analysis of cell interaction networks and their underlying molecular mechanisms. As a result, spatial transcriptomics serves as a uniquely advantageous tool in various fields of biomedical research.
Numerous technologies have been developed to capture transcriptomes with spatial resolution. Many of these methods have been commercialized and found utility in diverse applications, including the assessment of cell functionality in tissues, tracking organ development, analyzing tumor microenvironments, and unraveling pathology and disease mechanisms (Figure 2). This emphasizes the immense potential of spatial transcriptomics technologies in a wide range of biomedical areas, offering transformative opportunities for significant advancements in the field1.

Figure 2: Applications of spatial transcriptomics in cancer research
Despite the progress, substantial challenges persist. The applicability of most spatial transcriptome technologies is restricted to fresh or frozen specimens, rendering them incompatible with formalin-fixed paraffin-embedded (FFPE) samples, which are particularly prevalent in clinical contexts. Furthermore, among those technologies that are compatible with FFPE samples, many rely on targeted probe capture methods, limiting their ability to gather information beyond predetermined sequences within the coding regions. This limitation hinders comprehensive analyses, such as the investigation of gene fusions and alternative splicing, and the exploration of non-coding RNA - all of which are pivotal in fields such as pathology, neurology, oncology, and immunology2-4.
M20 Spatial: Exploring the Frontiers of Spatial Transcriptomics
Addressing the limitations encountered with current methodologies, M20 Genomics has leveraged its extensive expertise in random-primer technology to pioneer the world's first spatial whole transcriptome technology based on random primers: M20 Spatial.
Our pioneering technology offers a range of distinctive features:
1.Versatile Sample Compatibility: M20 Spatial offers unparalleled flexibility, efficiently processing a variety of sample types, including fresh, frozen, and FFPE specimens.
2.Unbiased Whole-Transcriptome Access: With M20 Spatial, the capture of the entire transcriptome is achieved, allowing for the thorough investigation of non-coding RNAs, including long non-coding RNAs (lncRNAs), thus broadening the scope of in-depth insights.
3.Comprehensive Full-Length Sequences: M20 Spatial excels in capturing the information of full-length transcripts, enabling the detailed examination of transcriptomic complexities such as alternative splicing, and enhancing the accuracy of analysis.
M20 Spatial heralds a transformative era in spatial transcriptomics, providing unmatched versatility, expansive coverage, and unparalleled depth of analysis. Our technology demonstrates exceptional proficiency unraveling intricate spatially resolved gene expression patterns and regulatory mechanisms, with outstanding performance.
M20 Spatial: Excelling Through Challenges with Exceptional Performance
M20 Spatial achieves a remarkable breakthrough by its applicability to formalin-fixed paraffin-embedded (FFPE) samples. As hospitals and biorepositories curate extensive archives of FFPE samples, numbering in the billions, these specimens represent an invaluable source to gain pathological insights. However, FFPE samples pose significant challenges for transcriptomic platforms due to nucleic acid degradation induced by formalin cross-linking and high-temperature paraffin infiltration during the preservation process. The degradation is further exacerbated by factors such as preservation duration and environmental conditions. As a consequence of the low RNA quality in FFPE samples, the availability of transcriptomic platforms compatible with such specimens remains highly constrained.
Despite these challenges, our comprehensive evaluation of FFPE samples from various human and mouse tissues has consistently demonstrated the exceptional performance of our M20 Spatial technology. We are thrilled to showcase high-quality spatial transcriptome data obtained from diverse mouse FFPE samples, including heart, brain and kidney sections, highlighting the outstanding capabilities of M20 Spatial.
Achieving Remarkable Sensitivity in Spatial Transcriptome Profiling
In a FFPE-preserved tissue section of a mouse heart, M20 Spatial exhibited impressive sensitivity. Across 3,048 barcoded spots, each with a diameter of 50 µm, a total of 31,072 genes were detected. The median gene count per spot stood at 3,651 (Figure 3), showcasing the excellent performance of M20 Spatial in capturing transcriptomic data.
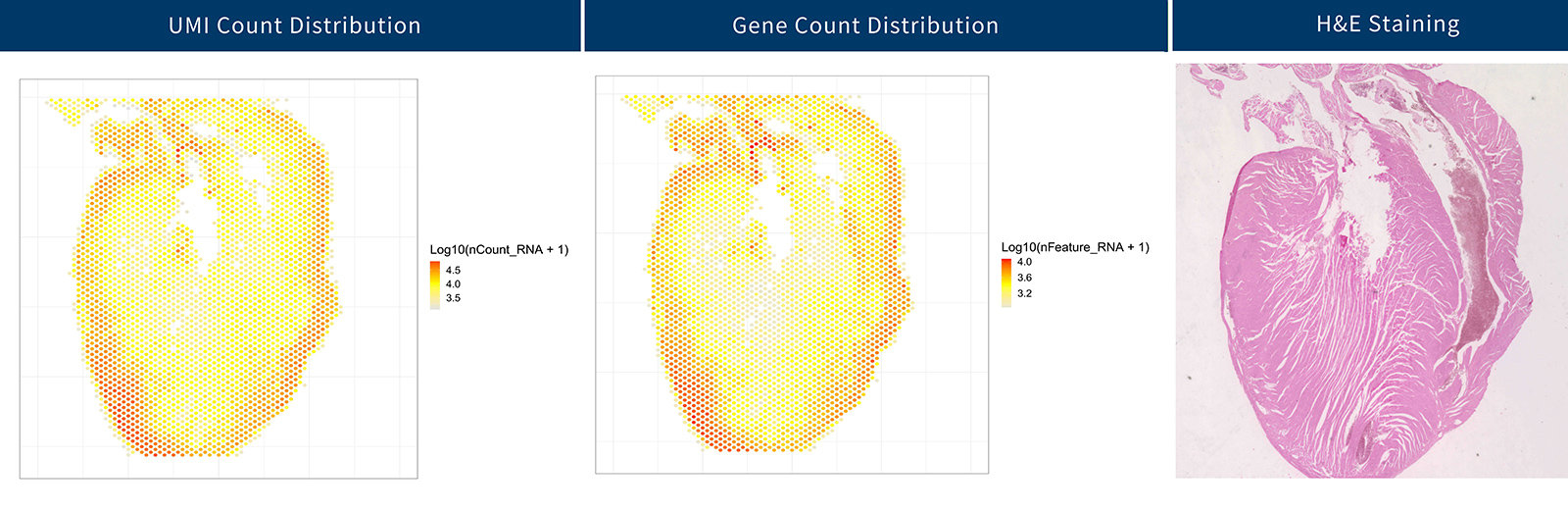
Figure 3: UMI and gene count distribution detected in sections of a FFPE mouse heart using M20 Spatial
The notable performance was further confirmed in a FFPE mouse brain tissue section. M20 Spatial successfully captured a total of 29,380 genes from 3,190 barcoded spots, resulting in a median gene count of 3,449 per spot (Figure 4).

Figure 4: UMI and gene count distribution detected in sections of a FFPE mouse brain using M20 Spatial
Unveiling Heterogeneity with Spatial Resolution
In the comprehensive analysis of FFPE samples sourced from diverse mouse organs our M20 Spatial technology showcased remarkable proficiency in differentiating diverse cell subpopulations. Leveraging UMAP clustering on transcriptomic data of mouse heart, brain and kidney sections, M20 Spatial accurately identifies multiple subpopulations within each sample (Figures 5-7, left panels).

Figure 5: UMAP clustering (left) and spatial projection (right) of cell subpopulations detected in a FFPE mouse heart section with M20 Spatial
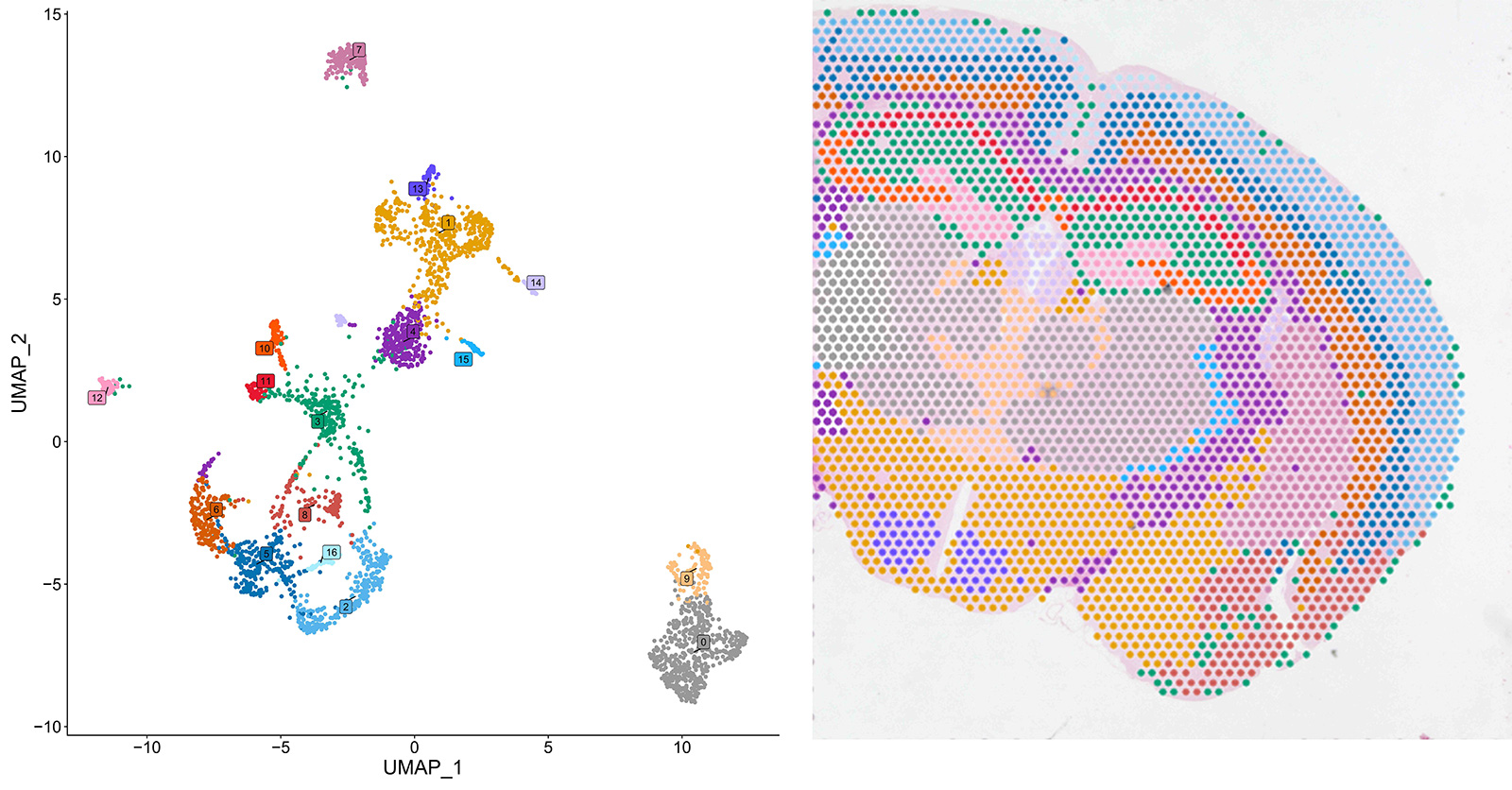
Figure 6: UMAP clustering (left) and spatial projection (right) of cell subpopulations detected in a FFPE mouse brain section with M20 Spatial
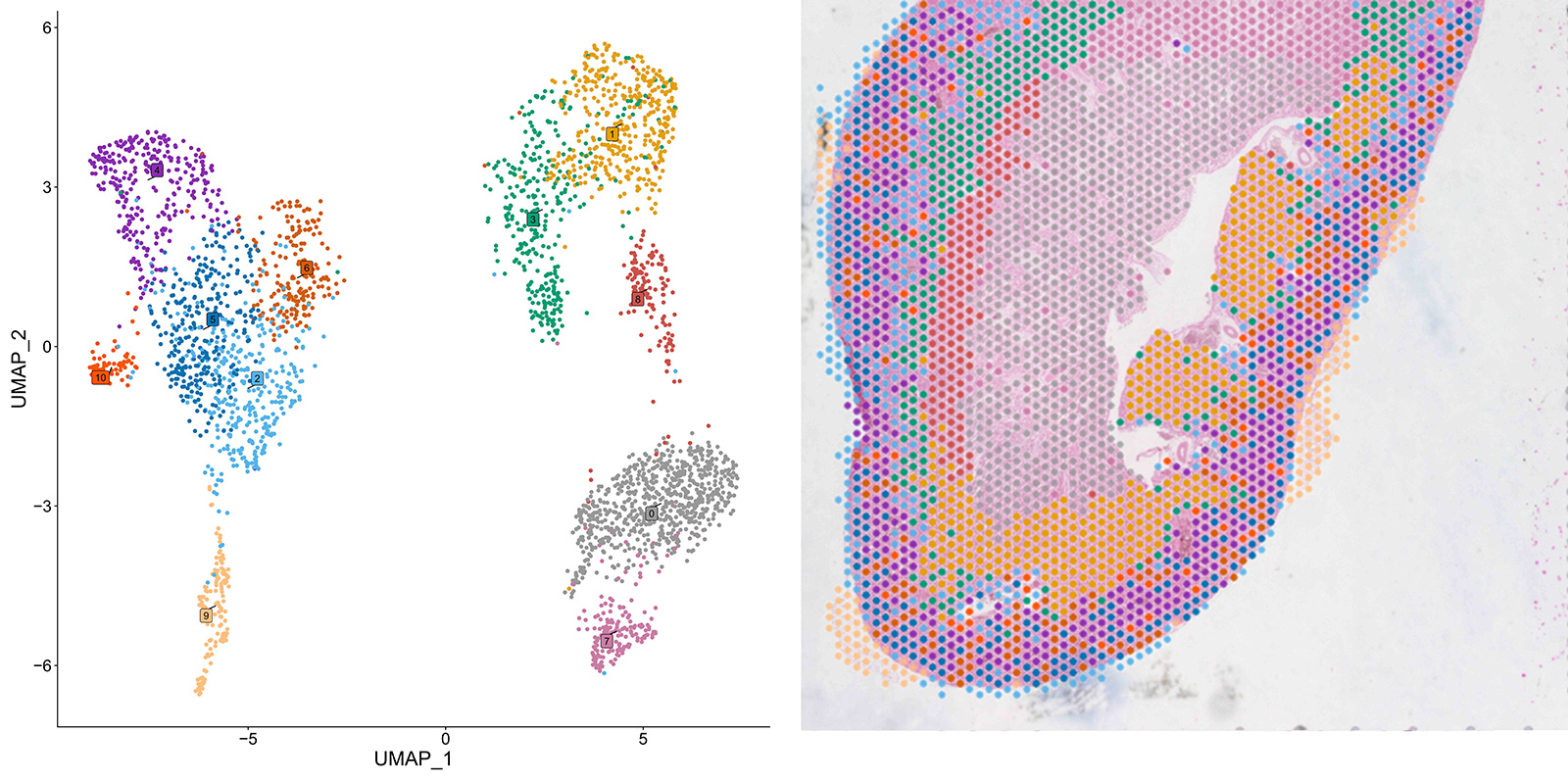
Figure 7: UMAP clustering (left) and spatial projection (right) of cell subpopulations detected in a FFPE mouse kidney section with M20 Spatial
The spatial projection of the identified subpopulations to their respective locations on the sample section reveals their correspondence to distinct tissue types within the organ (Figures 5-7, right panels). Notably, in complex organs like the brain, where cell types exhibit significant variation across different regions, the spatial projection of the subpopulations identified with M20 Spatial directly reflect the spatial organization of distinct brain regions (Figure 6).
These findings underscore the capability of M20 Spatial to accurately distinguish cellular subpopulations exhibiting heterogeneity at both spatial and transcriptomic levels, rendering it highly suitable for thorough exploration of spatial heterogeneity among cells within tissues and organs.
Full Coverage of Transcripts without Bias
Most spatial transcriptome technologies rely on poly(A) or targeted probes for RNA capture, often exhibiting strong preferences for specific transcript regions. This inherent bias leads to an overrepresentation of the 3' end or targeted sequences in the obtained transcriptome data.
In contrast, M20 Spatial transforms spatial transcriptomics by employing random primers. Our technology captures complete gene bodies from the 5' to 3' end uniformly (Figure 8), ensuring an accurate representation of the transcriptome.

Figure 8: mRNA 5'-3' coverage in M20 Spatial dataUnlocking Spatial Information on lncRNAs
Our random-primer-based technology demonstrates remarkable performance and allows to exceed previous limitations in spatial transcriptome sequencing. Most technologies currently enable the detection of protein-coding transcripts. However, M20 Spatial facilitates the detection of the entire transcriptome, including transcripts lacking poly(A) tails or exhibiting low-abundance.
Within the diverse group of non-coding RNAs, long non-coding RNAs (lncRNAs) emerge as key regulators across a multitude of biological processes, spanning from cancer development and immunity to stress responses and neural activities. Their substantial impact on cell physiology has pushed lncRNAs into the focus of scientific research in recent years3-4.
M20 Spatial effectively captures lncRNAs in FFPE sections from various mouse organs including heart, brain and kidney, representing 2-10% of the total detected RNAs (Figures 9-11). This unique capability not only underscores the versatility of M20 Spatial but also promises to unlock new avenues for groundbreaking discoveries in spatial biology, revolutionizing our understanding of cellular dynamics and disease mechanisms.
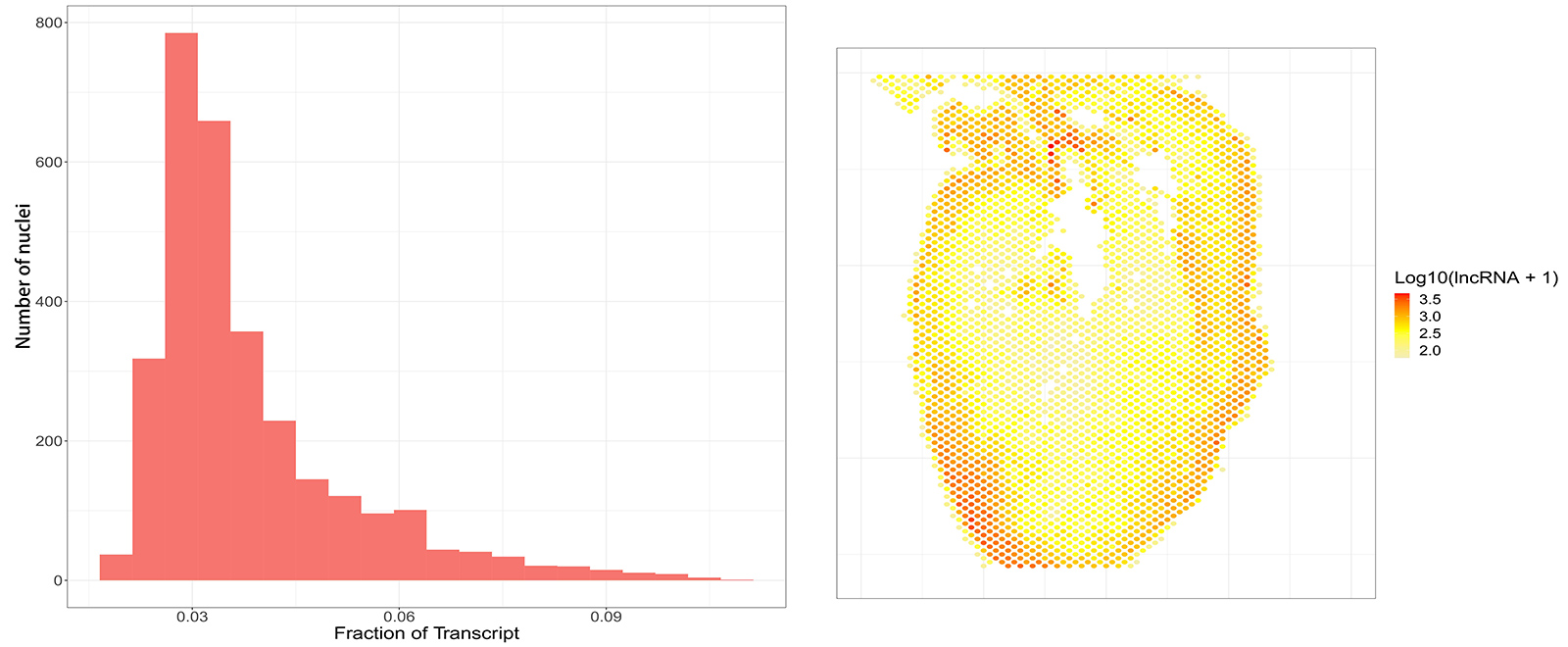
Figure 9: Percentage of lncRNA detected in a FFPE mouse heart section using M20 Spatial

Figure 10: Percentage of lncRNA detected in a FFPE mouse brain section using M20 Spatial

Figure 11: Percentage of lncRNA detected in a FFPE mouse kidney section using M20 Spatial
Our innovative spatial whole transcriptome technology, M20 Spatial, distinguishes itself by not only facilitating spatial transcriptome analysis across diverse sample types but also by delivering unparalleled, intricately comprehensive spatial transcriptome data. This remarkable capability represents a significant leap forward, promising to greatly expand the scope and precision of spatial transcriptomics applications in fields such as neuroscience, pathology, oncology, immunology, and beyond.
M20 Spatial heralds the beginning of a new era of research excellence, as our cutting-edge technology offers enhanced capabilities poised to redefine the boundaries of spatial transcriptomics. Based on this technology, we are thrilled to announce the forthcoming launch of our next-generation spatial transcriptome platform: the NATA series. Stay tuned for more updates!
We eagerly anticipate the release of M20 Spatial and its associated products to revolutionize research in the field. M20 Genomics remains dedicated to advancing scientific discovery through continuous innovation. With our commitment to pushing the boundaries of spatial transcriptomics, we aim to empower researchers worldwide to unlock new insights and drive progress in life sciences.
References
[1] Park HE, et al. Spatial Transcriptomics: Technical Aspects of Recent Developments and Their Applications in Neuroscience and Cancer Research. Adv Sci (Weinh). 2023 Jun;10(16):e2206939.
[2] Ma Y, et al. Non-coding RNA in tumor-infiltrating regulatory T cells formation and associated immunotherapy. Front Immunol. 2023 Aug;14:1228331.
[3] Ali SA, et al. The non-coding RNA interactome in joint health and disease. Nat Rev Rheumatol. 2021 Nov;17(11):692-705.
[4] Soutschek M, Schratt G. Non-coding RNA in the wiring and remodeling of neural circuits. Neuron. 2023 Jul;111(14):2140-2154.



