- Q What is the underlying principle of M20 Seq technology?A
The underlying principle of M20 Seq is the use of random primers, which allows for the comprehensive capture of full transcriptomes within single cells. This method effectively captures a diverse range of RNA species, including messenger RNAs (mRNAs) and non-coding RNAs such as long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs). By encompassing a broad spectrum of RNA biotypes, M20 Seq technology empowers researchers to perform multidimensional analyses at the single-cell level, revealing complex molecular landscapes with exceptional precision.

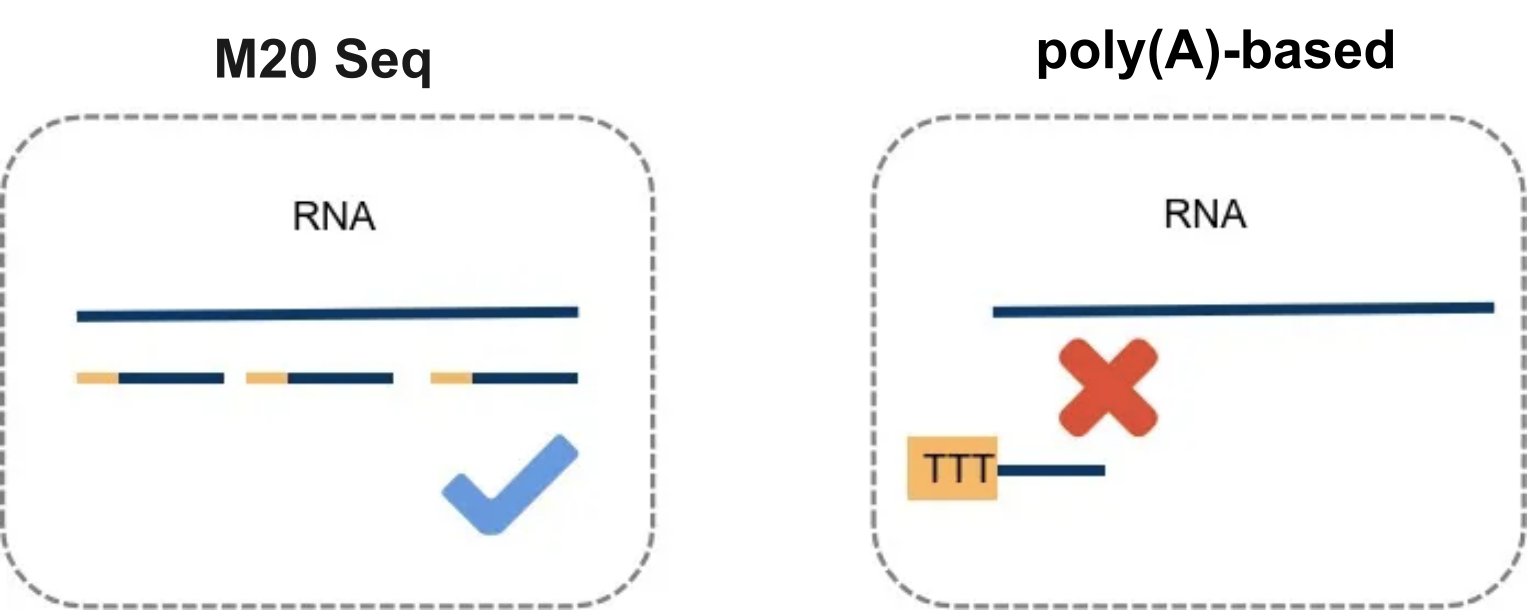
- Q Why does M20 Seq excel in single-cell RNA sequencing across challenging samples such as formalin-fixed and paraffin-embedded (FFPE), RNA-degraded, and bacterial samples?A
The M20 Seq technology employs random primers for RNA capture, making it highly effective in single-cell RNA sequencing of challenging sample types. Unlike traditional methods that rely on oligod(T) primers to capture RNA, M20 Seq does not require poly(A) tails. This feature ensures efficient RNA capture from various complex samples, including those with cross-linked RNAs in FFPE samples, partially degraded RNAs lacking poly(A) tail in certain samples, and bacterial samples, which naturally lack poly(A) tails.
- Q What are the components of the VITA Single-Cell Full-Length Transcriptome Platform?A
The VITA Single-Cell Full-Length Transcriptome Platform comprises the VITApilote Library Preparation Kits, the VITAcruizer Single-Cell Partitioning System, and the VITAseer Bioinformatics Software. Together, these components provide an end-to-end solution that empowers researchers to unlock the full potential of single-cell transcriptome analysis.
- Q What is the experimental workflow of the VITA Single-Cell Full-Length Transcriptome Platform?A
The VITA workflow begins with a suspension of single-nucleus, single-cell, or single-bacteria samples. This streamlined process utilizes the VITApilot Library Preparation Kits and the VITA Single-Cell Partitioning System to construct transcriptome libraries. Please note that certain sample types, such as FFPE, require an adapted workflow. For detailed information, please refer to the user guides.

- Q How long does the entire experimental process take, and at which stages can the experiment be paused?A
The full experimental workflow — from the preparation of single-cell/nucleus suspension to library construction — takes 1.5 to 2.5 days. Designed for operational flexibility, the process includes several checkpoints: after nucleus extraction, reverse transcription, cDNA extension, and cDNA amplification. These points offer high flexibility to pause the experiment, allowing effective time management without risking the quality of results.
- Q Is there a risk of nucleic acid materials being released from the cells or nuclei during the reverse transcription step?A
The risk of nucleic acid contamination during reverse transcription is effectively mitigated by the prior fixation step in the workflow. The fixation step involves 4% paraformaldehyde fixation immediately after sample collection. During this process, the formaldehyde forms methylene bridges with amino groups and protein structures, creating a three-dimensional network that entraps small biomolecules and ensures that nucleic acids remain confined within the cell or nucleus.
- Q How is single-cell barcoding achieved without relying on poly(A) capture?A
Single-cell barcoding without poly(A) capture is achieved through a ligation-based approach. After reverse transcription, an adapter is ligated to the cDNA. This adapter then hybridizes with sequences on barcoded beads, each carrying a unique cell barcode for every single cell or nucleus. During the extension process, the cell barcode sequence is incorporated into the cDNA fragments, ensuring accurate single-cell barcoding.

- Q What are the performance specifications of the instrument, the VITAcruizer Single-Cell Partitioning System?A
The equipment demonstrates robust capabilities across all parameters:

These specifications highlight the equipment’s versatility and efficiency, making it suitable for a wide array of biological research applications.
- Q Is the data generated with the VITA Single-Cell Full-Length Transcriptome Platform compatible with other single-cell analysis software?A
The initial data processing requires VITAseer Bioinformatics Software. Once processed, the data can be seamlessly integrated with commonly used downstream single-cell sequencing analysis tools, ensuring smooth and effective analysis.



